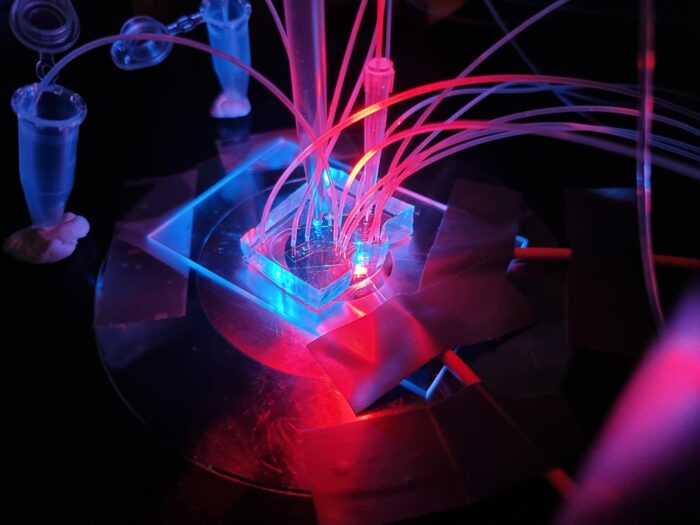
Welcome to the Laboratory of Microfluidics and Single Cell Biology at the University of Warsaw! We develop novel microfluidics tools for ultrahigh-throughput screening and single-cell genomics.

Single cell genomics
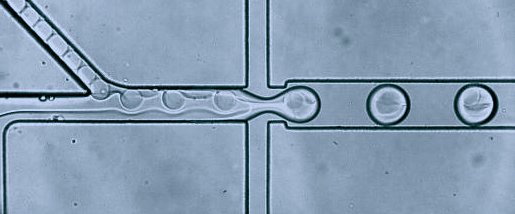
Organisms are built of a large number of diverse and highly specialized cells. To better understand the roles of each cell, we should study them at the single-cell level. One of the most significant achievements of analytical sciences in the last ten years was implementing a droplet microfluidic technology for analyzing single cells using RNA sequencing (RNA-seq). Droplet-based profiling of single-cell transcriptomes allowed for a very detailed description of the physiological state of thousands of single cells and led to an efficient determination of cell types, trajectories, and interactions. These methods are especially indispensable in research fields dealing with highly heterogeneous tissues – such as oncology, immunology, embryology, and other areas of biomedical research. In our research projects, we are applying already established unique techniques, as well as developing novel multi-step microdroplet assays to study transcriptomes and genomes of single cells.
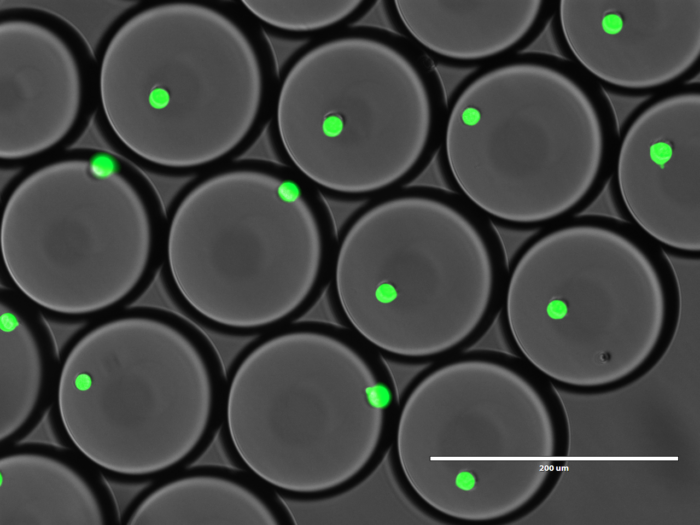
Ultrahigh-throughput screening in biology
We develop and implement ultrahigh-throughput technologies that will allow us to select the most efficient microbial consortia from thousands or even millions of combinations of environmental strains or microbial sub-populations. Droplet microfluidics offers an unprecedented increase in the number of reactions (up to 100 million), that can be measured within a single day using optical readouts, such as fluorescence or absorbance. The detection and sorting of droplets can be based on the bacterial growth or presence of detectable products of bioconversion. The most active strains selected using microfluidic ultrahigh throughput methods are then characterized using genomic and phenotypic analysis.